BENNERD
- Overview
- BENNERD DEMO
- CORD-NERD Data Set
- BENNERD Description
- Evaluation
- Acknowledgement
- Contact
- Citation
Table of contents generated with markdown-toc
Overview
In response to the coronavirus disease 2019 (COVID-19) for global research community to apply recent advances in NLP, COVID-19 Open Research Dataset (CORD-19) is proposed as a research challenge. The dataset consists of a resource of over 181,000 scholarly articles that are related to the infectious disease COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). To facilitate COVID-19 studies, since named entity recognition (NER) is considered a fundamental step in text mining system, data mining group of CS@UIUC has created CORD-NER dataset with comprehensive NE annotations. The annotations are based on distant or weak supervision. The CORD-NER dataset includes 29,500 documents from the CORD-19 corpus.
We present our recent efforts related to the coronavirus disease in 2019. Our main objective is to provide our recent biomedical entity linking system for end-users to facilitate and accelerate COVID-19 studies. We provide an web interface so that end-users can easily analyze their texts with our system. The evaluation of our method are also reported on this page with comparing with several existing state-of-the-art systems. We also release our UMLS-based automatically annotated CORD-NERD dataset and manually annotated test set that are used to evaluate our system to support the NLP community in developing the natural language processing (NLP) systems for the coronavirus disease.
BENNERD DEMO
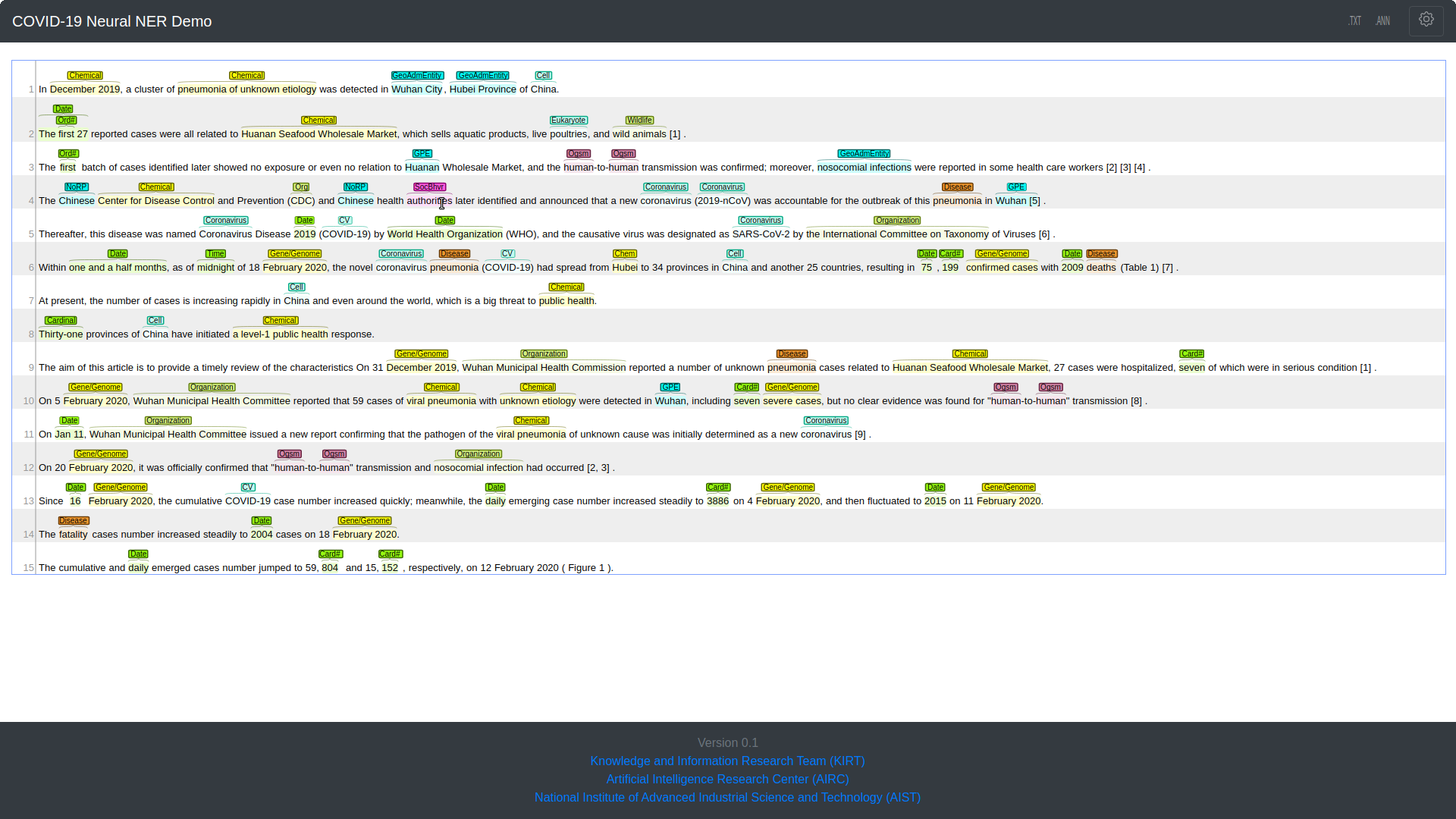
The BENNERD system provides a web interface to facilitate the process of text annotation and its disambiguation without any training for end users like researchers or biologists. In our demo, users can input a plain text of any biomedical document to extract entities with their corresponding types and UMLS CUI(s) in BRAT format. It also provides a feature to filter out some of the specific entities by their types via a simple checkbox list (the setting button on the top right of page).
Users can access our demo via http://prm-ezcatdb.cbrc.jp/bennerd/.
CORD-NERD Data Set
We are the first to perform entity linking (EL) task on CORD-19 data set. CORD-NER dataset comprises only NER task. To solve the EL task, we expand this dataset by leveraging a CUI for each mention in the CORD-NER dataset, we call this covid-19 open research dataset for named entity recognition and disambiguation CORD-NERD dataset that includes UMLS-based training, development, and test sets. Available at,
We use the most recent UMLS version 2020AA release that includes coronavirus-related concepts. To create a dataset for EL, we use a dictionary matching approach based on exact match using UMLS KB. CORD-NER includes 10,470,248 mentions, among which 6,794,126 and 3,676,122 mentions are respectively present and absent in the UMLS. Therefore, the entity coverage ratio of CORD-NER over the UMLS is 64.89%. We annotate the entity mentions that are not found in the UMLS with CUI_LESS. Finally we augment the CORD-NER dataset with CUI for each correspondence mention.
EL system is judged on two test sets: 1. CORD-NERD Test set or UMLS-based test set and 2. Manually annotated test set. These data sets are available from the following links.
- UMLS-based Test Set or CORD-NERD Test set
- Manually Annotated Test Set
It is worth to mention that both UMLS-based Test Set and Manually Annotated Test Set or even CORD-NERD dataset are based on the orginal offsets of CORD-NER dataset.
UMLS-based Test Set
To evaluate the EL performance on CORD-NERD, 302,166 mentions are assigned for 5,000 test set. We call this UMLS-based test set from CORD-NERD dataset.
Manually Annotated Test Set
In addition with the UMLS-based test set, we assigned a biologist to annotate 1,000 random sentences based on chemical, disease, and gene types along with its corresponding CUI to create a manually annotated test set.
Sample Data Format of Extended CORD-NER
Examples of annotation for an entity (T), a normalization (N) are shown in the following. Text-bound annotation identifies a specific span of text and assigns it a type. In text-bound annotation (T1) of a span “Angiotensin-converting enzyme 2”, 0 denotes start-offset and 31 denotes end-offset of the annotation span, where type is GENE_OR_GENOME. The normalization annotation (N1) is attached to the text-bound annotation (T1) which is associated with the unified medical language system (UMLS) entry with the UMLS concept unique identifier (CUI) as C0960880.
T1 GENE_OR_GENOME 0 31 Angiotensin-converting enzyme 2
N1 Reference T1 UMLS:C0960880
T2 GENE_OR_GENOME 33 37 ACE2
N2 Reference T2 UMLS:C1422064
T3 CORONAVIRUS 44 54 SARS-CoV-2
N3 Reference T3 UMLS:C5203676
T4 CHEMICAL 55 63 receptor
N4 Reference T4 UMLS:C0597357
T5 CORONAVIRUS 120 130 SARS-CoV-2
N5 Reference T3 UMLS:C5203676
T6 EVOLUTION 158 170 phylogenetic
N6 Reference T6 UMLS:cui_less
T7 WILDLIFE 195 198 bat
N7 Reference T7 UMLS:C1412726
T8 CORONAVIRUS 214 224 SARS-CoV-2
N8 Reference T3 UMLS:C5203676
T9 NORP 259 277 intermediate hosts
N9 Reference T9 UMLS:cui_less
T10 CORONAVIRUS 282 292 SARS-CoV-2
N10 Reference T3 UMLS:C5203676
BENNERD Description
Introduction
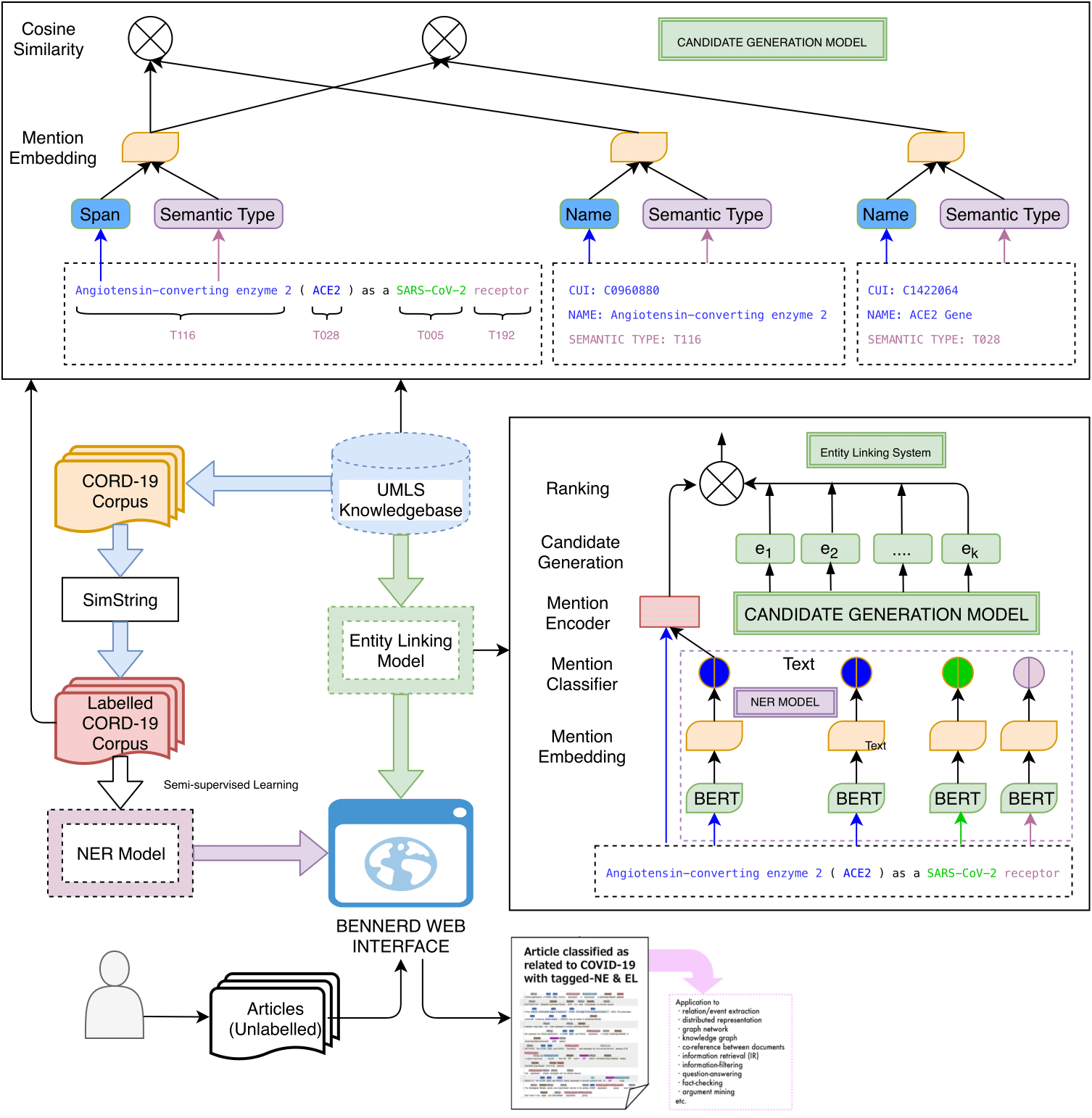
In this work, we present a BERT-based Exhaustive Neural Named Entity Recognition and Disambiguation (BENNERD) system by addressing CORD-NER data set. The entity disambiguation (ED) or entity normalization (EN) is a.k.a entity linking (EL) task. The objective of this work is to facilitate recent pandemic of corona virus disease 2019 (COVID-19) research that mainly covers biomedical domain, especially new entity types (e.g., coronavirus, viral proteins, and immune responses) by addressing CORD-NER dataset. In contrast to biomedical domain, it also gives a shed on general domain entity types (e.g., person, organization, location, date, group etc.). Moreover, along with flat or non-overlapping, the BENNERD system also solves nested or overlapping entities. The BENNERD system is composed of four models: NER that enumerates exhaustively all possible spans as potential entity mentions and classifies them into entity types, masked language model as BERT, candidate generation model to find a list of candidate entities, and candidate ranking model to disambiguate the entity for concept indexing.
Usage of BENNERD
Annotation tasks like entity mention detection and entity normalization based on CORD-NER can be performed in BENNERD for COVID-19 research. The CORD-NER contains 63 biomedical and general entity types therefore BENNERD can be applied on other biomedical corpora like GENIA, BC5CDR etc. In contrast to biomedical domain, BENNERD can be applied on general domain for mention detection and normalization in some extent as the unified medical language system (UMLS) is used as only knowledge base. BENNERD can be used to visualize annotation output for manual analysis and evaluation performances.
BENNERD System
BENNERD system mainly comprises two platforms: BENNERD web interface and BENNERD back-end server. The overall workflow of the BENNERD system is illustrated as below.
BENNERD Web Interface
In BENNERD web interface, the user interface contains input panel, load a sample tab, annotation tab, gear box tab, and .TXT and .ANN tabs. For a given text from users or loading a sample text from a sample list, the annotation tab will show the annotations with the text based on best NER- and EL-based training model. Different colors represent different entity types and, when the cursor floats over a coloured box representing an entity above text, the corresponding concept unique identifier (CUI) on the UMLS is shown. Users can save the machine readable text and annotation files as .txt and .ann where the .ann annotation file provides standoff annotation output in brat format.
BENNERD Back-end
The BENNERD back-end is for storing tools (e.g., NER, EL) that transform into a pipeline. It is composed of BERT-based exhaustive neural named entity recognition (NER) model and the prediction of NER model then fed to entity linking (EL) model that disambiguate the predicted entities by addressing candidate generation model (to find a list of candidate entities) and candidate ranking model.
Neural Named Entity Recognition
Named entity recognition (NER) is a task of finding entities with specific semantic types such as Protein, Cell, and RNA in text. We build neural NER model, based on the BERT model. The layer receives subword sequences and assigns contextual representations to the subwords via BERT. We generate mention candidates based on the same idea as the span-based model.
Entity Linking
The CORD-NER dataset gives a shed on entity recognition system, but it does not address entity linking (EL) task which is important to address COVID-19 research. For example, the mention SARS-CoV-2 needs to be disambiguated. Since the term SARS-CoV-2 in this sentence refers to a virus, it should be linked to an entry of a virus in the knowledge base (KB), not to an entry of ‘SARS-CoV-2 vaccination’, which corresponds to therapeutic or preventive procedure to prevent a disease. To address EL, we implement candidate generation model to find a list of candidate entities in the UMLS KB for linking and candidate ranking model to disambiguate the entity for concept indexing.
Summary
We presented the BENNERD system for entity linking, hoping that we can bring insights for the COVID-19 studies on making scientific discoveries. The BENNERD system is continually evolving; we will continue to improve the system as well as to implement new functions such as relation extraction to further facilitate COVID-19 research.
Evaluation
NER Performances
We evaluate our NER system from several aspects: comparison with existing state-of-the-art models, different pre-trained bert models, categorical performance, and evaluation on manually annotated test sets.
BENNERD Performance Comparison with Existing Models on CORD-NER Dataset
Since the manually annotated CORD-NER test set is not publicly available, we can not directly compare our system performances on the gene, chemical, and disease entity types. Instead, here we show the performance of gene, chemical, and disease based on our 5,000 test set.
------------------------------------------------------------------------------------------------------------------------
Model Gene Chemical Disease All
------------------------ ------------------------ ------------------------ ------------------------
Precision Recall F1(%) Precision Recall F1(%) Precision Recall F1(%) Precision Recall F1(%)
--------------- ------------------------ ------------------------ ------------------------- -------------------------
SciSpacy* 26.18 19.23 22.17 30.76 7.35 11.86 17.71 2.78 4.80 26.90 11.35 15.96
(BIONLP13CG)
--------------- ------------------------ ------------------------ ------------------------- -------------------------
SciSpacy* - - - 35.00 10.81 16.52 37.54 42.68 39.94 36.38 18.60 24.62
(BC5CDR)
--------------- --------- ------ ----- --------- ------ ----- --------- ------ ----- --------- ------ -----
BENNERD 76.07 74.80 75.45 83.55 84.60 84.07 84.85 84.90 84.92 80.85 80.81 80.83
-------------------------------------------------------------------------------------------------------------------------
- SciSpacy (BIONLP13CG) is a SciSpacy NER model trained on the BIONLP13CG corpus.
- SciSpacy (BC5CDR) is a SciSpacy NER model trained on the BC5CDR corpus.
- *Note: Our BENNERD results used ClinicalCovid BERT.
BENNERD NER Performances using Different Pre-trained BERT Models
Since our NER model is mainly based on BERT architecture and there are numerious pre-trained BERT models on different corpora, we tested the performance of BENNERD NER on some of the well-known pre-trained BERT models for the biomedical domain. We also applied the same training setting to make sure that the results are comparable. We judge the NER performnces of BENNERD on development- and test-sets using ClinicalCovid BERT, SciBERT, and Covid BERT Base. In development- and test-sets, we obtain the best performances 85.52% and 83.03% respectively in terms of F1-score using ClinicalCovid BERT.
--------------------------------------------------------------------------------
Pre-trained BERT
Model Development-set Test-set
---------------------------- ----------------------------
Precision Recall F1(%) Precision Recall F1(%)
------------------ --------- ------ -------- --------- ------ --------
ClinicalCovid BERT 84.62 86.43 85.52 82.83 83.23 83.03
SciBERT 84.03 87.05 85.51 82.16 83.81 82.98
Covid BERT Base 78.31 66.80 72.10 77.44 66.80 71.73
--------------------------------------------------------------------------------
- ClinicalCovid BERT is a BERT model trained on scientific corpora, namely ClinicalBERT and BioBERT.
- SciBERT is trained on papers from the corpus of semanticscholar.org.
- Covid BERT Base is based on fine-tuned a BERT base model on the CORD-19 corpus.
Categorical Performances Based on All Categories
We show the categorical performances of NER model trained on ClinicalCovid BERT over the CORD-NER dataset. Along with categorical performances on precision, recall, and F1-score, we also show the numbers of #TPs can be denoted as number of true positive outcomes where the model correctly preditcs the positive category, #Preds as number of predicted entities, and #Golds as number of gold entites of each category.
----------------------------------------------------------------------------------------------
Label Precision Recall F1-score(%) #TPs #Preds #Golds
---------------------------------- --------- ------ ----------- ------ ------- ------
ANATOMICAL_STRUCTURE 76.62 76.62 76.62 390 509 509
ARCHAEON 96.30 96.30 96.30 52 54 54
BACTERIUM 79.80 90.51 84.82 391 490 432
BODY_PART_ORGAN_OR_ORGAN_COMPONENT 87.03 84.79 85.90 3517 4041 4148
BODY_SUBSTANCE 95.56 96.72 96.14 1357 1420 1403
CARDINAL 82.36 90.94 86.44 54376 66021 59793
CELL 82.24 87.52 84.80 15278 18578 17456
CELL_COMPONENT 83.03 81.41 82.21 9789 11790 12024
CELL_FUNCTION 95.33 97.66 96.48 1797 1885 1840
CELL_OR_MOLECULAR_DYSFUNCTION 98.91 98.91 98.91 2267 2292 2292
CHEMICAL 83.55 84.60 84.07 94459 113056 111650
CORONAVIRUS 98.46 98.94 98.70 21928 22271 22162
DAILY_OR_RECREATIONAL_ACTIVITY 97.20 98.85 98.02 1460 1502 147
DATE 80.24 83.79 81.98 30231 37675 36080
DIAGNOSTIC_PROCEDURE 94.72 96.57 95.64 592 625 613
DISEASE_OR_SYNDROME 84.85 84.99 84.92 30717 36202 36140
EDUCATIONAL_ACTIVITY 90.91 93.75 92.31 30 33 32
EUKARYOTE 89.68 95.04 92.28 5268 5874 5543
EVENT 40.79 17.71 24.70 31 76 175
EVOLUTION 97.16 98.46 97.80 5301 5456 5384
EXPERIMENTAL_MODEL_OF_DISEASE 96.05 97.33 96.69 146 152 150
FAC 42.75 10.87 17.34 56 131 515
FOOD 89.12 93.40 91.21 467 524 500
GENE_OR_GENOME 76.07 74.83 75.45 71833 94426 95993
GOVERNMENTAL_OR_REGULATORY_ACTIVITY 65.76 99.18 79.08 121 184 122
GPE 78.40 74.71 76.51 7020 8954 9396
GROUP 98.48 99.25 98.86 11104 11275 11188
GROUP_ATTRIBUTE 88.10 100.0 93.67 74 84 74
HUMAN-CAUSED_PHENOMENON_OR_PROCESS 95.88 98.94 97.38 93 97 94
IMMUNE_RESPONSE 97.29 99.42 98.35 1725 1773 1735
INDIVIDUAL_BEHAVIOR 84.50 93.09 88.59 229 271 246
INJURY_OR_POISONING 93.60 95.12 94.35 117 125 123
LABORATORY_OR_TEST_RESULT 95.63 97.47 96.54 810 847 831
LABORATORY_PROCEDURE 93.24 95.43 94.32 4030 4322 4223
LANGUAGE 78.57 61.11 68.75 22 28 36
LAW 43.98 22.51 29.78 95 216 422
LIVESTOCK 96.67 97.26 96.96 2091 2163 2150
LOC 70.63 43.34 53.72 267 378 616
MACHINE_ACTIVITY 97.35 96.08 96.71 147 151 153
MATERIAL 94.80 90.46 92.58 237 250 262
MOLECULAR_FUNCTION 96.31 97.91 97.10 4930 5119 5035
MONEY 50.97 32.99 40.05 158 310 479
NORP 70.48 70.79 70.63 1294 1836 1828
ORDINAL 85.55 95.82 90.39 4220 4933 4404
ORG 66.33 50.37 57.26 9289 14004 18440
ORGANISM 85.80 83.32 84.54 9015 10507 10820
ORGAN_OR_TISSUE_FUNCTION 89.84 94.01 91.88 345 384 367
PERCENT 66.33 42.76 52.00 65 98 152
PERSON 56.02 51.16 53.48 4027 7188 7872
PHYSICAL_SCIENCE 96.80 93.08 94.90 121 125 130
PRODUCT 52.60 35.28 42.23 930 1768 2636
QUANTITY 71.15 73.38 72.25 3095 4350 4218
RESEARCH_ACTIVITY 98.63 98.75 98.69 2600 2636 2633
SIGN_OR_SYMPTOM 95.07 96.39 95.73 1310 1378 1359
SOCIAL_BEHAVIOR 95.73 98.18 96.94 538 562 548
SUBSTRATE 95.99 98.46 97.21 1341 1397 1362
THERAPEUTIC_OR_PREVENTIVE_PROCEDURE 97.28 98.86 98.06 7551 7762 7638
TIME 86.67 85.33 86.00 1437 1658 1684
TISSUE 78.10 74.66 76.34 4223 5407 5656
VIRAL_PROTEIN 89.39 91.09 90.23 1053 1178 1156
VIRUS 89.18 95.09 92.04 5383 6036 5661
WILDLIFE 98.43 97.56 97.99 1438 1461 1474
WORK_OF_ART 34.75 14.44 20.40 41 118 284
----------------------------------------------------------------------------------------------
Entity Linking Performances
We evaluate our candidate EL system performances based on two settings. In the setting 1, we train the CUIs based on manually annotated MedMention dataset. In the setting 2, the BENNERD model is trained on automatically annotated CORD-NERD dataset.
Entity Linking Performances of BENNERD on UMLS-based Test Set
We show the EL performances on the UMLS-based test set. We report Accuracy@n, where n = 1, 10, 20, 30, 40, 50. Accuracy@1, gold candidate was ranked highest. Accuracy@{10, 20, 30, 40, 50} indicates, gold candidate was in top 10, 20, 30, 40 or in 50 predictions of the candidate ranker. In the performances of BENNERD with NER’s prediction, we fed the NER’s predictions to the entity linking model to judge the entity linking performances. In contrast to the performances of BENNERD with gold NEs and NER’s true positive entities, we fed the gold entities and true positive entities (where the model correctly predicts the positive category) respectively to judge the entity linking performances.
-------------------------------------------------------------------------------------------
Model UMLS-based Test Set
----------------------------------------------------------
A@1 A@10 A@20 A@30 A@40 A@50(%)
----------------------------- -------- -------- -------- -------- -------- --------
Setting1: Trained on Manually Annotated MedMention Dataset
-------------------------------------------------------------------------------------------
BENNERD + Predicted NEs 27.61 44.56 49.74 51.88 53.08 54.19
BENNERD + Gold NEs 29.78 48.33 53.89 56.22 57.53 58.74
BENNERD + True Positive NEs 30.31 48.91 54.60 56.95 58.27 59.49
-------------------------------------------------------------------------------------------
Setting2: Trained on UMLS-based Automatically Annotated Extended CORD-NER Dataset
-------------------------------------------------------------------------------------------
BENNERD + Predicted NEs 47.46 64.32 67.70 69.87 71.12 72.07
BENNERD + Gold NEs 50.73 69.31 73.10 75.58 77.03 78.13
BENNERD + True Positive NEs 53.90 73.06 76.90 79.36 80.79 81.87
-------------------------------------------------------------------------------------------
Entity Linking Performances of BENNERD on Manually Annotated Test Set
We show the EL performances on the manually annotated test set. On the manually annotated test set, the performances in terms of Accuracy@{1, 10, 20, 30, 40, 50} are based on NER’s predictions that fed to entity linking model.
-------------------------------------------------------------------------------------------
Model Manually Annotated Test Set
---------------------------------------------------------
A@1 A@10 A@20 A@30 A@40 A@50(%)
------------------------- ------- ------- ------- ------- ------- -------
BENNERD + Setting1 24.27 42.95 47.07 48.81 50.00 50.92
BENNERD + Setting2 31.84 50.25 54.53 56.87 58.39 60.12
BENNERD + String Matching 30.21 41.00 41.00 41.00 41.00 41.00
-------------------------------------------------------------------------------------------
Acknowledgement
This work is based on results obtained from a project commissioned by the Public/Private R&D Investment Strategic Expansion PrograM (PRISM)
Contact
- Mohammad Golam Sohrab: sohrab.mohammad@aist.go.jp
- Khoa N. A. Duong
- Hai-Long Trieu
- Makoto Miwa
- Goran Topić
- Masami Ikeda
- Hiroya Takamura
Citation
If you find this work helpful, please use the following citation:
@inproceedings{bennerd_2020,
title = "BENNERD: A Neural Named Entity Linking System for COVID-19",
author = "Sohrab, Mohammad Golam and Duong, Khoa and Miwa, Makoto and Topić, Goran and Masami, Ikeda and Takamura, Hiroya",
booktitle = "Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations",
month = oct,
year = "2020",
address = "Online",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/2020.emnlp-demos.24",
pages = "182--188",
}
Thanks!!!